 |
 |
 |
 |
 |
Our research is primarily focussed on the transport properties of narrow graphene nanoribbons. We apply the theoretical formalism based on density functional theory combined with single-particle Green's function technique to numerically evaluate the transmission probabilities of nanoscale junctions. Electron transmission of a graphene nanoribbon on which a molecule is adsorbed shows molecular fingerprints of Fano resonances, i.e., characteristic features of frontier orbitals and conformations of physisorbed molecules.
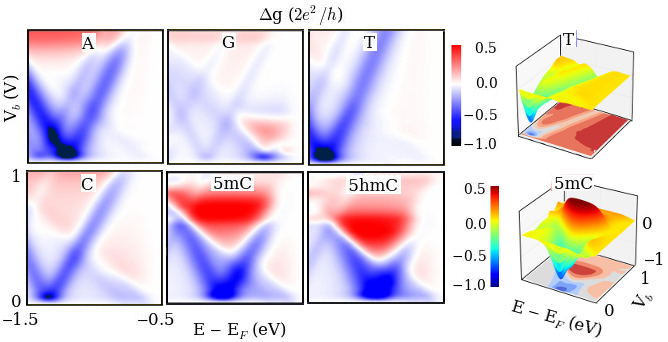
Utilizing these resonance profiles, we demonstrate two-dimensional molecular electronics spectroscopy (2D MES). The differential conductance with respect to bias and gate voltages not only distinguishes different types of nucleobases (A, G, T, C) for DNA sequencing but also recognizes methylated nucleobases (5mC, 5hmC) which could be related to cancerous cell growth. This 2D MES could open an exciting field to recognize single molecule signatures at atomic resolution. The advantages of the 2D MES over the one-dimensional (1D) current analysis can be comparable to those of 2D NMR over 1D NMR analysis.
The 2D MES should be useful for single molecule spectroscopy at atomic resolution based on Fano resonance driven molecular fingerprinting and conformational analysis, while widely used laser driven molecular spectroscopy suffers from low resolution due to the diffraction limit of light wave. This single molecule recognition can be further utilized to distinguish different types of nucleobases for fast DNA sequencing without highly time-consuming complicated chemical/optical labeling procedure. It should be noted that some nucleobases in DNA are frequently present in methylated form. The most dramatic consequence of DNA methylation would be cancer. Hence, recognition of methylated bases could give a precancer warning sign. Since methylation alters the energy levels and coupling strengths of MOs, the 2D MES utilizing Fano resonance should be able to distinguish methylated nucleobases from normal nucleobases. Hence, this method would have a great advantage over the available methylated DNA detection such as bisulfite treatment and amplification.
